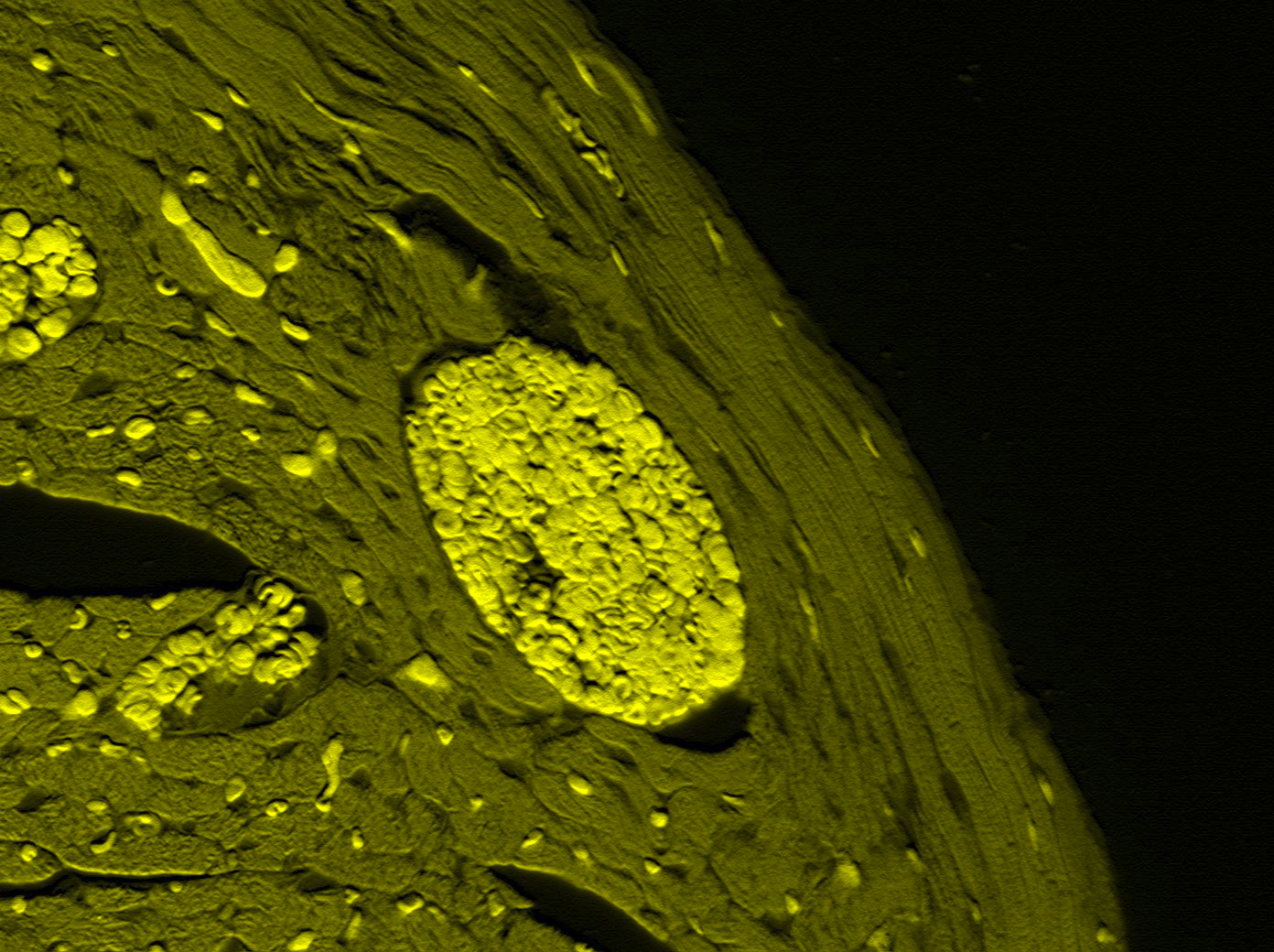
Biochemistry & MCB.
The final aim at this area of BIFI is to understand and to control biological systems depending on proteins that have interest for chemical, biotechnological, pharmacological and biomedical applications. Knowledge of protein behaviour at molecular and cellular levels allows interpretation of the macroscopic mechanisms of the cellular functions in which they are involved, but many of the parameters controlling these processes still remain unknown. Proteins adopt an organized three-dimensional structure closely related with their function that can be regulated by the interaction with other biomolecules and/or small organic molecules. Defective structural arrangements can prevent protein interaction with other molecules, provoking many different illnesses in human beings.
Many other illnesses are also produced by infectious viruses and microorganisms, and a way of stopping them could be to block a step of their vital cycle involving a protein or biomolecule. Moreover, small molecules from the environment can also be either toxic inhibitors or activatiors of particular activities in different organisms, and in many cases they might be used as effectors of the expression some genes, and the production or action of particular proteins. The research lines in Biochemistry and Molecular and Cellular Biology at BIFI study different biological systems involved in key metabolic routes that serve as model for other systems combining classic methodologies from this area with biophysical and computational methods. Applications of the obtained knowledge are additionally being used to control and to modulate the behaviour of particular systems with benefits for the society.
- Genetic regulation and physiology of cyanobacteria
- Plant evolutionary and genetic biology
- Functional genomics of the OXPHOS system (GENOXPHOS)
- Development of antimicrobials and mechanisms of resistance (D2AMR)
- Apoptosis and metabolism
- Computational genomics and systems BioMedicine
Genetic regulation and physiology of cyanobacteria
Head of the Research Line:
María F. Fillat Castejón
Researchers:
Teresa Bes Fustero
Emma Sevilla Miguel
Ana Alonso Simón
Jorge Guío Martínez
Irene Oliván Muro
Inés Federío Zalaya
SUMMARY OF THE RESEARCH LINE
Cyanobacteria are microorganisms that perform oxygenic photosynthesis and are capable of colonizing the most extreme habitats. Due to their abundance and ubiquity, cyanobacteria play a key role in the carbon and nitrogen cycles and constitute the base of the food chain in aquatic ecosystems. Some species of cyanobacteria are capable of fixing atmospheric nitrogen and have been used in the production of fertilizers. Likewise, the use of cyanobacteria for the production of biodiesel or the removal of heavy metals from wastewater are areas of work in continuous development.
The research line studies the regulation of iron metabolism in cyanobacteria and its relationship with nitrogen metabolism, oxidative stress, cyanotoxin production and biofilm formation. All these processes are interrelated through a family of transcriptional regulators called FUR (ferric uptake regulator). Most cyanobacteria express three FUR paralogs called FurA (Fur), Zur (FurB), and PerR (FurC). Although the best studied facet of these proteins is that of transcriptional regulator, in cyanobacteria they have a multifunctional character, acting through various strategies that are not well characterized, complementing their activity as transcriptional regulators.
On the other hand, the research’s group work has shown that FUR proteins are global regulators in cyanobacteria, which in turn control other regulators, two-component systems and sigma factors. Therefore, work is being done on to identication of new regulatory networks mediated by FUR proteins, characterizing a series of regulators modulated by FUR paralogs whose function is unknown.
Another of the group’s fields of study is the formation of cyanobacteria biofilms, due to the environmental implications and applications that these formations have. For this, the nitrogen-fixing filamentous cyanobacterium Anabaena sp. PCC7120 is being used as a model.
The study of the potential of cyanobacteria in bioremediation, specifically their ability to biodegrade lindane or γ-hexachlorocyclohexane (γ-HCH) and its isomers (α-,β-,δ-HCH) has revealed that overexpression mutants of FurC in Anabaena PCC7120, have a greater bioremediation capacity. Contamination by hexachlorocyclohexane is a worldwide problem, which very directly affects the Autonomous Community of Aragon due to the activity that the lindane producing industry, Inquinosa, carried out in Sabiñánigo a few years ago and because the discharges it generated are still stored in Aragonese territory. The studies carried out by the group in recent years have allowed us to advance in the understanding of the regulation of the expression of genes involved in the degradation of lindane and its isomers, and to identify a candidate gene for the design of a biosensor. This biosensor could allow early warning of risk in the event that these organochlorines enter bodies of water. In this context, work will be done on i) the development of a prototype whole-cell HCH biosensor, and ii) the identification of intermediates of the lindane degradation pathway in cyanobacteria.
The main objectives of the research group are:
– Identification of regulatory networks mediated by the FUR family in cyanobacteria, and characterization of new transcriptional regulators that are part of these networks and have unknown functions.
– Study of the formation of biofilms in Anabaena: conditions that facilitate their formation, genes involved and potential uses of biofilms in bioremediation and biofertilization.
– Characterization of the Fur regulator of the pathogen Clostridium difficile and evaluation of this protein as a potential therapeutic target by altering its activity by screening chemical libraries.
-Application of cyanobacteria as relevant organisms in bioremediation. Specifically, the ability of the model cyanobacterium Anabaena PCC7120 to degrade lindane and its isomers is being studied. These studies have led to obtain information that can be used to develop a biosensor for early warning of the presence of lindane in waters.
Relevant publications
1.-Unbalancing Zur (FurB)-mediated homeostasis in Anabaena sp. PCC7120: Consequences on metal trafficking, heterocyst development and biofilm formation. Olivan-Muro I, Sarasa-Buisan C, Guio J, Arenas J, Sevilla E, Fillat MF. Environ Microbiol. 2023 Jun 14. doi: 10.1111/1462-2920.16434.
2. Responses of Anabaena sp. PCC7120 to lindane: Physiological effects and differential expression of potential lin genes. Guío J, Fillat MF, Peleato ML, Sevilla E. Microbiology open. 2023 Jun;12(3):e1355. doi: 10.1002/mbo3.1355.
3. Expanding the FurC (PerR) regulon in Anabaena (Nostoc) sp. PCC 7120: Genome-wide identification of novel direct targets uncovers FurC participation in central carbon metabolism regulation. Sarasa-Buisan C, Guío J, Peleato ML, Fillat MF, Sevilla E. PLoS One. 2023 Aug 7;18(8):e0289761. doi: 10.1371/journal.pone.0289761. eCollection 2023.
4. Metal binding and oligomerization properties of FurC (PerR) from Anabaena sp. PCC7120: an additional layer of regulation? Sarasa-Buisan C, Emonot E, Martínez-Júlvez M, Sevilla E, Velázquez-Campoy A, Crouzy S, Bes MT, Michaud-Soret I, Fillat MF. Metallomics. 2022 Oct 20;14(10):mfac077. doi: 10.1093/mtomcs/mfac077.
5. FurC (PerR) from Anabaena sp. PCC7120: a versatile transcriptional regulator engaged in the regulatory network of heterocyst development and nitrogen fixation. Sarasa-Buisan C, Guio J, Broset E, Peleato ML, Fillat MF, Sevilla E. Environ Microbiol. 2022 Feb;24(2):566-582. doi: 10.1111/1462-2920.15552.
6. Thioredoxin Dependent Changes in the Redox States of FurA from Anabaena sp. PCC 7120. Guío J, Bes MT, Balsera M, Calvo-Begueria L, Sevilla E, Peleato ML, Fillat MF. Antioxidants (Basel). 2021 Jun 4;10(6):913. doi: 10.3390/antiox10060913.
7. Fur-like proteins: beyond the Fur uptake regulator (Fur) paralog. Sevilla E, Bes MT, Peleato Ml, Fillat MF. Archives in Biochemistry and Biophysics,701: 108770. doi: 10.1016/j.abb.2021.108770.
8. 2-oxoglutarate modulates the affinity of FurA for the ntcA promoter in Anabaena sp. PCC 7120. Guio, Jorge; Sarasa-Buisan, Cristina; Velazquez-Campoy, Adrian; et ál. FEBS Lett. 2020 Jan;594(2):278-289. doi: 10.1002/1873-3468.13610.
9. Regulation by FurC in Anabaena Links the Oxidative Stress Response to Photosynthetic Metabolism. Sevilla E, Sarasa-Buisan C, González A, Cases R, Kufryk G, Peleato ML, Fillat MF. Plant Cell Physiol. 2019 Aug 1;60(8):1778-1789. doi: 10.1093/pcp/pcz094.2.
10. Redox-Based Transcriptional Regulation in Prokaryotes: Revisiting Model Mechanisms. Sevilla E, Bes MT, González A, Peleato ML, Fillat MF. Antioxid Redox Signal. 2019 May 1;30(13):1651-1696. doi: 10.1089/ars.2017.7442. Epub 2018 Sep 18.
11. Transcriptional regulators: valuable targets for novel antibacterial strategies. González A, Fillat MF, Lanas Á. Future Med Chem. 2018 Mar 1;10(5):541-560. doi: 10.4155/fmc-2017-0181. Epub 2018 Feb 20. Review.
12. Molecular basis for the integration of environmental signals by FurB from Anabaena sp. PCC 7120. Sein-Echaluce VC, Pallarés MC, Lostao A, Yruela I, Velázquez-Campoy A, Luisa Peleato M, Fillat MF. Biochem J. 2018 Jan 5;475(1):151-168. doi: 10.1042/BCJ20170692.
Main research projects
1.- E35_23R: Grupo de referencia biología estructural. Gobierno de Aragón. Duración: 01/01/2023 – 31/12/2025. Subvención: 37.743 euros. PI: Marta Martínez-Júlvez y Mª Teresa Bes Fustero. Investigadores: 10.
2.- Redes reguladoras implicadas en la respuesta a estrés y la formación de biofilms en cianobacterias. Identificación de nuevas rutas vinculadas a las proteínas FUR. IP María Francisca Fillat Castejón. Agencia Estatal de Investigación. 01/06/2020 – 31/05/2024. Duración: 4 años
3.- Multifuncionalidad de las proteínas FUR en cianobacterias: mecanismos alternativos de regulación del metabolismo y contribución a la formación de biofilms. MINECO. Duración: 01/01/2017 – 31/12/2019. Subvención: 140.000 euros. PI: María F. Fillat Castejón. Investigadores: 5.
4.- B18 BIOLOGÍA ESTRUCTURAL. Gobierno de Aragón. Duración: 01/01/2014 – 31/12/2016. Subvención: 20.609 euros. PI: María Luisa Peleato Sánchez. Investigadores: 16.
5.- Identificación y aplicaciones sintéticas de nuevos biocatalizadores oxidativos en biotecnología industrial. (BIOXCAT). Ministerio de Ciencia e Innovación. Duración: 01/01/2022-31/12/2023. Subvención: 190.900 euros. PI: Patricia Ferreira y Juan Mangas. Investigadores: 5.
Collaborators
- Dr. I. Michaud-Soret (Institut de Recherche en Technologie et Science pour le Vivant. CEA, Grenoble, Francia)
- Dr. R. Helm (Virginia Tech, USA)
- Dr. Himadri Pakrasi (Washington St. University, USA)
- Conrad Mullineaux (Queen Mary, University of London, UK)
- Dr. I. Luque (Instituto de Bioquímica vegetal y Fotosíntesis, CSIC, Sevilla, España)
- Dr. Giulia Veronesi (ESFR, Grenoble, Francia)
- Dr. A. Lostao (Instituto de Nanociencia de Aragón, Universidad de Zaragoza, España)
- Dr. A. Lanas (Instituto Aragonés de Ciencias de la Salud, Zaragoza, España)
Contacto:
María F. Fillat, e-mail: fillat@unizar.es
X del grupo: @cyanofur
Plant Evolutionary and genetic Biology
Head of the Research Line:
Pilar Catalán Rodríguez
Researchers:
Pilar Catalán Rodríguez
Ernesto Pérez Collazos
Samira Ben-Menni Schuler
Maria Ángeles Decena Rodríguez
Miguel Campos Cáceres
Alba Sotomayor Alge
Yana Malymon
SUMMARY OF THE RESEARCH LINE
The general objective of our research is to establish reference model plant systems (Brachypodium) through genomic-functional and ecological analyses, and the potential transfer of the results obtained with them to other plants of agronomic and biofuel interest. In addition to this, we deepen the research of wild grasses in global hotspots, unraveling their genomes and their evolution, the interactions of grasses with their endophytic fungi and with the soil microbiome and their co-evolutionary and adaptive implications. We also study the genetic characterization and conservation of endemic or threatened plants.
Line 1: Comparative and functional genomics of model plants
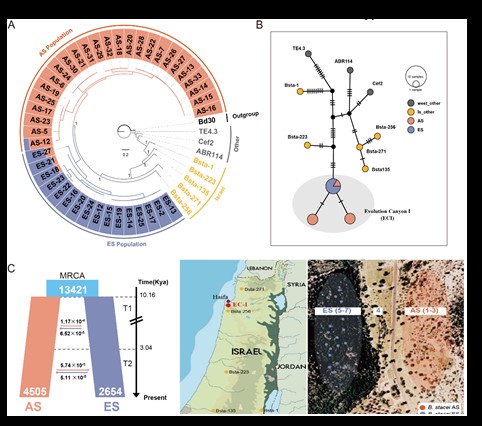 The specific objectives of our studies in Brachypodium model plants seek to obtain complete genomes and annotated pan-genomes of various key species, and to elucidate, through comparative functional genomics and phylogenomics, genomes, transcriptomes and elements involved in gene expression (factors of transcription, chromatin remodelers), the regulatory mechanisms associated with adaptations to abiotic stresses (drought, nutrients) and phenotypic changes in diploid/allopolyploid and annual/perennial species of Brachypodium. We also investigate the impact of transposons on the expression of diversity related to ecologically relevant phenotypic and functional traits, and the potential subgenomic overdominance and epigenomic regulation that affects differential gene expression after polyploidization, and during the development and environmental adaptation of these plants.
The specific objectives of our studies in Brachypodium model plants seek to obtain complete genomes and annotated pan-genomes of various key species, and to elucidate, through comparative functional genomics and phylogenomics, genomes, transcriptomes and elements involved in gene expression (factors of transcription, chromatin remodelers), the regulatory mechanisms associated with adaptations to abiotic stresses (drought, nutrients) and phenotypic changes in diploid/allopolyploid and annual/perennial species of Brachypodium. We also investigate the impact of transposons on the expression of diversity related to ecologically relevant phenotypic and functional traits, and the potential subgenomic overdominance and epigenomic regulation that affects differential gene expression after polyploidization, and during the development and environmental adaptation of these plants.
Line 2: Systematics and evolution of angiosperms
Using the genomic advances obtained in Brachypodium, we address the analysis of the barely explored genomes of pasture and forage grasses of high ecological and economic value (genus Festuca and subfamily Pooideae), through comparative and functional genomics approaches. In this context we also analyze the grass-endophyte interactions. To do this, we will also sequence genomes of fungal species of the genus Epichloë, investigating the phylogenomics of these haplo/heteroploid fungi, and their possible co-evolution with their host grasses. Additionally, we will investigate through metabolomic analyzes the benefits of mutualism in Festuca ecotypes whose endophytes provide them with adaptive and agronomic advantages, including the synthesis of alkaloids with insecticidal function, for their selection and possible registration as cultivars for the improvement of pastures, lawns and herbaceous covers.
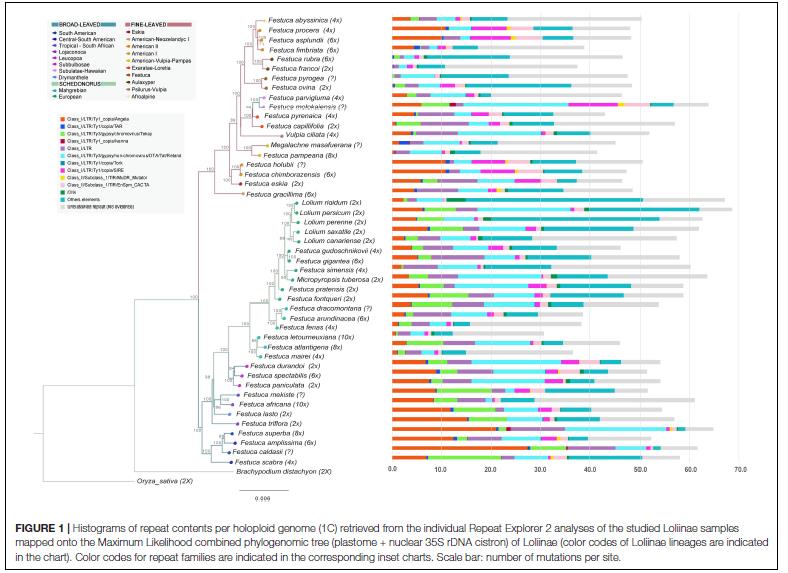
Line 3: Genetics, ecology and conservation of plants in Aragon and other regions
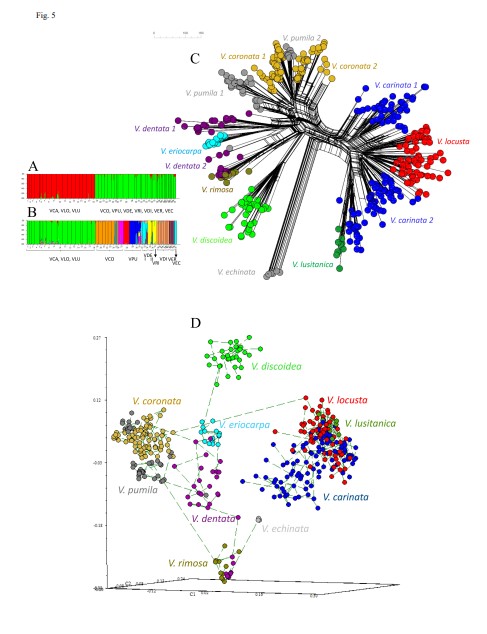 Our research strategy also includes our scientific and social commitment to genetic (and genomic) conservation studies of endemic and threatened plants, both in Aragon and other regions. To do this we will use herbarium collections, population genetics (and genomics) data, bio-/phylogeography and ecological niche modeling, as well as reproductive biology analysis and in-situ and ex-situ conservation techniques, and the dissemination of these results to society. Our contributions constitute a valuable tool for transferring basic research results to applied research in flora conservation that allows better management of species at risk of extinction and the monitoring and control of invasive species.
Our research strategy also includes our scientific and social commitment to genetic (and genomic) conservation studies of endemic and threatened plants, both in Aragon and other regions. To do this we will use herbarium collections, population genetics (and genomics) data, bio-/phylogeography and ecological niche modeling, as well as reproductive biology analysis and in-situ and ex-situ conservation techniques, and the dissemination of these results to society. Our contributions constitute a valuable tool for transferring basic research results to applied research in flora conservation that allows better management of species at risk of extinction and the monitoring and control of invasive species.
Relevant publications
1. Mu W, Li K, Yang Y, Breiman A, Yang J, Wu Y, Wu S, Zhu M, Liu J, Nevo E, Catalán P. 2023. Scattered differentiation of unlinked loci across the genome underlines ecological divergence of the selfing grass Brachypodium stacei. PNAS 120: e2304848120
2. Mu W, Li K, Yang Y, Breiman A, Yang J, Wu J, Zhu M, Wang S, Catalán P, Nevo E, Liu J. 2023. Subgenomic stability of progenitor genomes during repeated allotetraploid origins of the same grass Brachypodium hybridum. Molecular Biology Evolution (accepted).
3. Campos M, Kelley E, Gravendeel B, Médail F, Christenhusz JM, Fay MF, Catalán P, Leitch IJ, Forest F, Wilkin P, Viruel J. 2023. Genomic, spatial and morphometric data for discrimination of four species in the Mediterranean Tamus clade os yams (Dioscorea, Dioscoreaceae). Annals of Botany 131(4):635-654.
4. Scarlett V, Lovell J, Shao M, Phillips J, Shu S, Lusinska J, Goodstein D, Jenkins J, Grimwood J, Barry K, Chalhoub B, Schmutz J, Hasterok R, Catalan P, Vogel J. 2022. Multiple origins, one evolutionary trajectory: gradual evolution characterizes distinct lineages of allotetraploid Brachypodium. Genetics, 10.1093/genetics/iyac146.
5. Sancho R, Catalán P, Contreras-Moreira B, Juenger TE, Des Marais DL. 2022. Patterns of pan-genome occupancy and gene co-expression under water-deficit in Brachypodium distachyon. Molecular Ecology 31:5285-5306. doi: 10.1111/mec.16661
6. Moreno-Aguilar MF, Inda LA, Sánchez A, Arnelas I, Catalán P. 2022. Evolutionary dynamics of the repeatome explains contrasting differences in genome sizes and hybrid and polyploid origins of grass Loliinae lineages. Frontiers in Plant Sciences 13: 901733. doi: 10.3389/fpls.2022.901733
7. Hasterok R, Catalan P, Hazen SP, Roulin AC, Vogel JP, Wang K, Mur LAJ. 2022. Brachypodium: Twenty years as a grass biology model system; the way forward? Trends in Plant Science 27: 1002-1016. https://doi.org/10.1016/j.tplants.2022.04.008.
8. Sancho R, Inda LA, Diaz-Perez A, Des Marais D, Gordon SP, Vogel JP, Lusinska J, Hasterok R, Contreras-Moreira B, Catalán P. 2022. Tracking the ancestry of known and ‘ghost’ homeologous subgenomes in model grass Brachypodium polyploids. The Plant Journal 109(6):1535-1558. doi: 10.1111/tpj.15650. Epub 2022 Feb 8. PMID: 34951515.
9. Arnelas I, Pérez-Collazos E, López-Martínez J, Devesa JA, Catalán P. 2022. Molecular systematics of Valerianella Mill. (Caprifoliaceae): challenging the taxonomic value of genetically controlled carpological traits. Plants 11: 1276. https://doi.org/10.3390/plants11101276
10. Moreno-Aguilar MF, Inda LA, Sánchez A, Catalán P, Arnelas I. 2022. Phylogenomics and systematics of overlooked Mesoamerican and South American polyploid broad-leaved Festuca grasses differentiate F. sects. Glabricarpae and Ruprechtia and F. subgen. Asperifolia, Erosiflorae, Mallopetalon and Coironhuecu (subgen. nov.). Plants 11, 2303. doi.org/10.3390/plants11172303
Main research projects
1. Co-evolution and adaptive speciation grass-endophyte (Festuca, Brachypodium, Epichloë) in a pan-genomic framework (COEVOHOLOGENOME). Ministerio de Ciencia e Innovación. Proyecto PID2022-140074NB-I00. 2023- 2025. IP. P. Catalán. Investigadores participantes del grupo: E. Perez, R. Sancho, M.A. Decena, M. Campos, A. Sotomayor, Y. Malymon. 281.250,00 euros
2. Integrative genomic characterization of the Brachypodium polyploid model to unravel bases of success of polyploidy in flowering plants. Joint Genome Institute. Department of Energy. Government of United States of America. Community Science Program (CSP) proposal 503504. INRA-Evry, University of Silesia, University of Aberystwyth. 2018-2022. IP: P. Catalán. Investigadores participantes: E. Perez, R. Sancho, M.A. Decena, M. Campos, 7,5 TB of genomic and transcriptomic sequencing
3. Gramínea-endófito-microbioma: evolucion, ecología y mejora ecosistémica de pastos y cubiertas herbáceas. Ministerio de Ciencia e Innovación. Proyecto TED2021-131073B-I00. 2022-2024. IP P. Catalán. Investigadores participantes: E. Perez, R. Sancho, M.A. Decena, M. Campos, A. Sotomayor. 113.850,00 euros
4. Gramínea-Microbioma: registro de variedades protegidas de ecotipos de élite y sus microbiomas. Ministerio de Ciencia e Innovación. Proyecto PDC2022-133712-I00. 2022-2024. IP. P. Catalán. Investigadores participantes: E. Perez, R. Sancho, M. Campos, A. Sotomayor. 149.500,00 euros
Collaborators
- John Vogel. Joint Genome Institute. Walnut Creek CA USA.
- David Des Marais. Massachusetts Institute of Technology. Boston MA USA.
- Richard Amasino. University of Wisconsin-Madison. Madison WI USA.
- Bruno Contreras-Moreira. Estación Experimental de Aula Dei – CSIC. Zaragoza Spain.
- Isabel Marques. Universidade de Lisboa. Lisboa Portugal.
- Antonio Manzaneda. Universidad de Jaén. Jaén Spain.
- Robert Hasterok and Alexander Betekhtin. University of Silesia. Katowice Poland.
- Luis Mur and John Doonan. Aberystwyth University. Aberystwyth UK.
- Liliana Giussani. Instituto Botánico Darwinion – CONICET. Buenos Aires Argentina.
- Marina Olonova. Tomsk State University. Tomsk Russia.
- Teresa Garnatje. Instituto Botánico de Barcelona – CSIC. Barcelona Spain.
CONTACT
Functional genomics of the OXPHOS system (GENOXPHOS)
Head of the Research Line:
Patricio Fernández Silva
Pilar Bayona Bafaluy
Researchers:
Nuria Garrido Pérez
Patricia Meade Huerta
Raquel Moreno Loshuertos
SUMMARY OF THE RESEARCH LINE
- The activity of the line “Functional genomics of the OXPHOS system: GENOXPHOS”, included in the area of Biochemistry and Molecular Biology, is based on the understanding of the genetic and environmental factors that control the organization and remodeling of the mitochondrial OXPHOS system. More specifically, we want to analyze and understand the effect of supercomplex (SCs) assembly variants and mtDNA mutations on OXPHOS function and on processes such as tumorigenesis or the response to treatments with nanoparticles. Thus, our main lines of work are the following:- Evaluation of the effects of alterations in the apoptosis induction factor on aspects of mitochondrial function such as the production of reactive oxygen species, the organization of respiratory complexes and SCs, OXPHOS function, mitochondrial biogenesis or induction of cell death through the Parthanatos pathway.- Study of the role of mitochondria in the processes of tumorigenesis and metastasis, analyzing mitochondrial function and the organization of the OXPHOS system in tumor cell lines with different invasive capacity, as well as their response to treatment with metabolic drugs such as dichloroacetate, metformin or deoxyglucose, among others.- Analysis of the effect of increasing temperature on the assembly and stability of respiratory SCs and OXPHOS function.
– Generation and analysis of the operation of intracellular nanothermometers
– Development of an intracellular local magnetic hyperthermia system as antitumor therapy.
– Molecular study of mutations in genes encoded in the mtDNA or in the nuclear genome that affect the OXPHOS system.
– Obtaining reprogrammed cells (iPSCs) from fibroblasts to differentiate them into the most commonly affected tissues in patients.
Relevant publications
1.Moreno-Loshuertos, R., Movilla, N., Marco-Brualla, J., Soler-Agesta, R., Ferreira, P., Enríquez, J. A., & Fernández-Silva, P. A Mutation in Mouse MT-ATP6 Gene Induces Respiration Defects and Opposed Effects on the Cell Tumorigenic Phenotype. International Journal of Molecular Sciences, (2023), 24(2), 1300. https://doi.org/10.3390/ijms24021300
2. Moreno-Loshuertos, R., Marco-Brualla, J., Meade, P., Soler-Agesta, R., Enriquez, J. A., & Fernández-Silva, P. How hot can mitochondria be? Incubation at temperatures above 43 °C induces the degradation of respiratory complexes and supercomplexes in intact cells and isolated mitochondria. Mitochondrion, (2023), 69, 83–94. https://doi.org/10.1016/j.mito.2023.02.002.
3. Ruth Soler-Agesta, Joaquín Marco-Brualla, Patricio Fernández-Silva, Pilar Mozas, Alberto Anel, Raquel Moreno Loshuertos. Transmitochondrial Cybrid Generation Using Cancer Cell Lines. (JoVe) J. Vis. Exp. (2023), (193), e65186, doi:10.3791/65186 (2023). URL: jove.com/video/65186.
4. Y Gu, R. Piñol, R. Moreno-Loshuertos, C. D. S. Brites, J. Zeler, A. Martínez, G. Maurin- Pasturel, P. Fernández-Silva, J. Marco-Brualla, P. Téllez, R. Cases, R. Navarro Belsué, D. Bonvin, L. D. Carlos*, and A. Millán* Local Temperature Increments and Induced Cell Death in Intracellular Magnetic Hyperthermia. ACS Nano 2023, 17, 6822–6832. DOI: 10.1021/acsnano.3c00388.
5. Soler-Agesta R, Marco-Brualla J, Minjárez-Sáenz M, Yim CY, Martínez-Júlvez M, Price MR, Moreno-Loshuertos R, Ames TD, Jimeno J, Anel A. PT-112 Induces Mitochondrial Stress and Immunogenic Cell Death, Targeting Tumor Cells with Mitochondrial Deficiencies. Cancers (Basel). (2022); 14(16), 3851. doi: 10.3390/cancers14163851.
6. Novo N, Romero-Tamayo S, Marcuello C, Boneta S, Blasco-Machin I, Velázquez- Campoy A, Villanueva R, Moreno-Loshuertos R, Lostao A, Medina M, Ferreira P. Beyond a platform protein for the degradosome assembly: The Apoptosis-Inducing Factor as an efficient nuclease involved in chromatinolysis. PNAS Nexus (2022), 2(2), 312. doi: 10.1093/pnasnexus/pgac312.
7. Jiménez-Salvador I, Meade P, Iglesias E, Bayona-Bafaluy P, Ruiz-Pesini E Developmental origins of Parkinson disease: Improving the rodent models..Ageing Res Rev. 2023 Apr;86:101880. doi: 10.1016/j.arr.2023.101880. Epub 2023 Feb 10.
8. Ruiz-Pesini E, Bayona-Bafaluy MP, Sanclemente T, Puzo J, Montoya J, Pacheu-Grau D. Mitochondrial Genetic Background May Impact Statins Side Effects and Atherosclerosis Development in Familial Hypercholesterolemia. Int J Mol Sci. 2022 Dec 28;24(1):471. doi: 10.3390/ijms24010471.
9. Bayona-Bafaluy MP, López-Gallardo E, Emperador S, Pacheu-Grau D, Montoya J, Ruiz-Pesini E. Is population frequency a useful criterion to assign pathogenicity to newly described mitochondrial DNA variants? Orphanet J Rare Dis. 2022 Aug 19;17(1):316. doi: 10.1186/s13023-022-02428-0.
10. Bayona-Bafaluy MP, Montoya J, Ruiz-Pesini E. Oxidative phosphorylation system and cell culture media. Trends Cell Biol. 2021 Aug;31(8):618-620. doi: 10.1016/j.tcb.2021.05.003. Epub 2021 May 26.
Main research projects
1.“Nanotermometro intracelular para el estudio termico de la fisiologia celular y terapia del cancer por hipertemia magnetica local:PID2021-124354NB-I00 : Ministerio Ciencia Innovación y Universidades; IPs Angel Millán (CSIC/)/Raquel Moreno Loshuertos (Universidad de Zaragoza). 01/09/2022 al 31/08/2025. 700,00 Euros
2. PID2021-124354NB-I00. Plan Estatal de Investigación Científica, Técnica y de Innovación. Ministerio Ciencia Innovación y Universidades; convocatoria 2021. IPs: Mª Pilar Bayona Bafaluy y Eduardo Ruiz Pesini (Universidad de Zaragoza): 01/09/2022 al 31/08/2025
Collaborators
- Dr. José Antonio Enriquez. Centro Nacional de Investigaciones Cardiovasculares (CNIC). Madrid-Spain
- Massimo Zeviani. MBU-MRC. Cambridge-UK
- Eva Monleón. Dtpo. de Anatomía e Histología Humanas- UZ
Development of Antimicrobials and Mechanisms of Resistance

Head of the Research Line:
José Antonio Aínsa Claver
Santiago Ramón-García
Researchers:
José Antonio Aínsa Claver, PI
Santiago Ramón-García, PI
Ainhoa Lucía Quintana, Postdoc
Clara Aguilar Pérez, PhD Student
Ernesto Anoz Carbonell, PhD Student
Ana Cristina Millán Placer, PhD Student
Marta María Gómara Lomero, PhD Student
María Pilar Arenaz Callao, PhD Student
Lara Muñoz Muñoz, PhD Student
Begoña Gracia Díaz, Laboratory Technician
SUMMARY OF THE RESEARCH LINE
Research line Development of Antimicrobials and Mechanisms of Resistance is committed to study mechanisms of resistance to antimicrobial agents in diverse microbial pathogens and to use this information for identifying novel molecules with antimicrobial activity and characterise their mechanisms of action and resistance. This work is funded by public grants got in competitive calls at the national and international level.
In recent years, we have characterised several efflux pumps from Mycobacterium tuberculosis and we have contributed to identify compounds that evading resistance mediated by efflux pumps have an increased antimicrobial activity. We are characterising novel drug targets, not only in Mycobacterium but also in other bacterial pathogens, and exploring novel molecules (peptides,…) as alternatives to conventional antibiotics.
During the last 7 years, we have published 15 scientific articles, we have got a patent related with the diagnostic value of a specific resistance gene and have made a number of communications to national and international conferences.
Our research is getting insight into novel perspectives, such as the use of nanoparticles for administering antibiotics, or combination of molecules with antimicrobial activity.
Relevant publications
1.- Discovery of antimicrobial compounds targeting bacterial type FAD synthetases. Sebastián M, Anoz-Carbonell E, Gracia B, Cossio P, Aínsa JA, Lans I, Medina M. J Enzyme Inhib Med Chem. 2018 Dec;33(1):241-254. doi: 10.1080/14756366.2017.1411910. PMID: 29258359
2.- The EU approved antimalarial pyronaridine shows antitubercular activity and synergy with rifampicin, targeting RNA polymerase. Mori G, Orena BS, Franch C, Mitchenall LA, Godbole AA, Rodrigues L, Aguilar-Pérez C, Zemanová J, Huszár S, Forbak M, Lane TR, Sabbah M, Deboosere N, Frita R, Vandeputte A, Hoffmann E, Russo R, Connell N, Veilleux C, Jha RK, Kumar P, Freundlich JS, Brodin P, Aínsa JA, Nagaraja V, Maxwell A, Mikušová K, Pasca MR, Ekins S. Tuberculosis (Edinb). 2018 Sep;112:98-109. doi: 10.1016/j.tube.2018.08.004. PMID: 30205975
3.- Synergy between Circular Bacteriocin AS-48 and Ethambutol against Mycobacterium tuberculosis. Aguilar-Pérez C, Gracia B, Rodrigues L, Vitoria A, Cebrián R, Deboosère N, Song OR, Brodin P, Maqueda M, Aínsa JA. Antimicrob Agents Chemother. 2018 Aug 27;62(9). pii: e00359-18. doi: 10.1128/AAC.00359-18. PMID: 29987141
4.- Boldine-Derived Alkaloids Inhibit the Activity of DNA Topoisomerase I and Growth of Mycobacterium tuberculosis. García MT, Carreño D, Tirado-Vélez JM, Ferrándiz MJ, Rodrigues L, Gracia B, Amblar M, Ainsa JA, de la Campa AG. Front Microbiol. 2018 Jul 24;9:1659. doi: 10.3389/fmicb.2018.01659. PMID: 30087665
5.- Total Synthesis of Ripostatin B and Structure-Activity Relationship Studies on Ripostatin Analogs. Glaus F, Dedić D, Tare P, Nagaraja V, Rodrigues L, Aínsa JA, Kunze J, Schneider G, Hartkoorn RC, Cole ST, Altmann KH. J Org Chem. 2018 Jul 6;83(13):7150-7172. doi: 10.1021/acs.joc.8b00193. PMID: 29542926
6.- New active formulations against M. tuberculosis: Bedaquiline encapsulation in lipid nanoparticles and chitosan nanocapsules. L.De Matteis, D.Jary, A.Lucía, S.García-Embid, I.Serrano-Sevilla, D.Pérez, J.A.Ainsa, F.P.Navarro, J.M. de la Fuente. Chemical Engineering Journal. Volume 340, 15 May 2018, Pages 181-191.
7.- Structure Guided Lead Generation toward Nonchiral M. tuberculosis Thymidylate Kinase Inhibitors. Song L, Merceron R, Gracia B, Quintana AL, Risseeuw MDP, Hulpia F, Cos P, Aínsa JA, Munier-Lehmann H, Savvides SN, Van Calenbergh S. J Med Chem. 2018 Apr 12;61(7):2753-2775. doi: 10.1021/acs.jmedchem.7b01570. PMID: 29510037
8.- Ionophore A23187 shows anti-tuberculosis activity and synergy with tebipenem. Huang W, Briffotaux J, Wang X, Liu L, Hao P, Cimino M, Buchieri MV, Namouchi A, Ainsa JA, Gicquel B. Tuberculosis (Edinb). 2017 Dec;107:111-118. doi: 10.1016/j.tube.2017.09.001. PMID: 29050757
9.- How can nanoparticles contribute to antituberculosis therapy? Costa-Gouveia J, Aínsa JA, Brodin P, Lucía A. Drug Discov Today. 2017 Mar;22(3):600-607. doi: 10.1016/j.drudis.2017.01.011. PMID: 28137645
10.- Antituberculosis drugs: reducing efflux=increasing activity. Rodrigues L, Parish T, Balganesh M, Ainsa JA. Drug Discov Today. 2017 Mar;22(3):592-599. doi: 10.1016/j.drudis.2017.01.002. PMID: 28089787
11.- Structure-Activity Relationships of Spectinamide Antituberculosis Agents: A Dissection of Ribosomal Inhibition and Native Efflux Avoidance Contributions. Liu J, Bruhn DF, Lee RB, Zheng Z, Janusic T, Scherbakov D, Scherman MS, Boshoff HI, Das S, Rakesh, Waidyarachchi SL, Brewer TA, Gracia B, Yang L, Bollinger J, Robertson GT, Meibohm B, Lenaerts AJ, Ainsa J, Böttger EC, Lee RE. ACS Infect Dis. 2017 Jan 13;3(1):72-88. doi: 10.1021/acsinfecdis.6b00158. PMID: 28081607
12.- Identification of Aminopyrimidine-Sulfonamides as Potent Modulators of Wag31-mediated Cell Elongation in Mycobacteria. Vinayak Singh, Neeraj Dhar, János Pató, Gaëlle S. Kolly, Jana Korduláková, Martin Forbak, Joanna C. Evans, Rita Székely, Jan Rybniker, Zuzana Palčeková, Júlia Zemanová, Isabella Santi, François Signorino-Gelo, Liliana Rodrigues, Anthony Vocat, Adrian S. Covarrubias, Monica G. Rengifo, Kai Johnsson, Sherry Mowbray, Joseph Buechler, Vincent Delorme, Priscille Brodin, Graham W. Knott, José A. Aínsa, Digby F. Warner, György Kéri, Katarína Mikušová, John D. McKinney, Stewart T. Cole, Valerie Mizrahi, Ruben C. Hartkoorn. Mol Microbiol. 2017 Jan;103(1):13-25. doi: 10.1111/mmi.13535. PMID: 27677649
13.- Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Bailo R, Bhatt A, Aínsa JA. Biochem Pharmacol. 2015 Aug 1;96(3):159-67. doi: 10.1016/j.bcp.2015.05.001. PMID: 25986884.
14.- Measuring efflux and permeability in mycobacteria. Rodrigues L, Viveiros M, Aínsa JA. Methods Mol Biol. 2015;1285:227-39. doi: 10.1007/978-1-4939-2450-9_13. PMID: 25779319.
15.- Spectinamides: a new class of semisynthetic antituberculosis agents that overcome native drug efflux. Lee RE, Hurdle JG, Liu J, Bruhn DF, Matt T, Scherman MS, Vaddady PK, Zheng Z, Qi J, Akbergenov R, Das S, Madhura DB, Rathi C, Trivedi A, Villellas C, Lee RB, Rakesh, Waidyarachchi SL, Sun D, McNeil MR, Ainsa JA, Boshoff HI, Gonzalez-Juarrero M, Meibohm B, Böttger EC, Lenaerts AJ. Nat Med. 2014 Feb;20(2):152-8. doi: 10.1038/nm.3458. PMID: 24464186.
Main research projects
1.- El fenotipo silente de Mycobacterium tuberculosis: persistencia y latencia. Ministerio de Economía y Competitividad. Universidad de Zaragoza. SAF2017-84839-C2-1-R. 01/01/2018 – 31/12/2020. IP José Antonio Aínsa Claver.
2.- Identification of novel therapies for difficult to treat cystic fibrosis pulmonary infections caused by mycobacteria using an innovative technology: synergy screens of clinically approved drugs. Unión Europea. Universidad de Zaragoza. 01/04/2018 – 31/03/2019. IP: Santiago Ramon Garcia.
3.- NAREB – Nanotherapeutics for antibiotic resistant emerging bacterial pathogens. Unión Europea. Universidad de Zaragoza. 01/02/2014 – 31/07/2018. IP: José Antonio Aínsa Claver.
4.- SAF-2013-48971-C2-2-R: Aplicaciones biomédicas de AS-48: una proteína con amplio espectro de actividad antimicrobiana. MINECO – Ministerio de Economia y Competitividad. Universidad de Zaragoza. 01/01/2014 – 31/07/2018. IP: José Antonio Aínsa Claver.
5.- MM4TB – More medicines for tuberculosis. Unión Europea. Universidad de Zaragoza. 01/02/2011 – 31/01/2016. IP: José Antonio Aínsa Claver.
Collaborators
- Adela G. De la Campa, Centro Nacional de Microbiología, Instituto de Salud Carlos III (Majadahonda, Madrid, Spain). Desarrollo de inhibidores frente a la topoisomerasa de tuberculosis.
- Mercedes Maqueda, Universidad de Granada (Granada, Spain). Estudio de actividad antimicrobiana de la bacteriocina AS-48.
- Serge van Calenbergh, Universidad de Gent (Gent, Belgium). Nuevos inhibidores de timidilato kinasa de tuberculosis.
- Concepción González-Bello (Universidad de Santiago de Compostela, Santiago de Compostela, Spain). Inhibidores de dehidroquinasas en tuberculosis.
Apoptosis and metabolism
Head of the Research Line:
José Alberto Carrodeguas Villar
Researchers:
Beatriz Sáenz de Buruaga/PhD
Laura Bueno Martínez/Student TFG
Sara García Gadea/Student TFG
SUMMARY OF THE RESEARCH LINE
Our work has focused in different lines during the last years, as stated next:
1- Studying PSAP/Mtch1 function (Presenilin 1-associated protein/mitochondrial carrier homolog 1). PSAP interacts with presenilin 1, part of the gamma secretase complex, involved in Alzheimer’s disease. It is also known as mitochondrial carrier homolog 1 (Mtch1), since it contains a domain conserved in outer mitochondrial membrane carriers, although it is located in the inner membrane. Mtch1 induces cell death upon overexpression in culture.
We have identified two isoforms generated by alternative splicing that localize into the inner membrane through inner targeting sequences and that contain two proapoptotic domains. Mtch1 can induce apoptosis in the absence of Bax and Bak, two proapoptotic members of the Bcl-2 family. It does not appear to have a role in transport, but it could function as a receptor for yet-to-be-identified ligands in the surface of mitochondria.
We have studied the Drosophila knockout of mtch1 and we are preparing a publication in this respect, in collaboration with researchers from the Biomedical Research Institute, CSIC-UAM, Madrid.
2- Bcl-2 proteins. Proteins from this family are essential for the fulfillment of cell death, integrating several cellular signals. Their major known roles depend on protein-protein interactions for the induction or the inhibition of initial steps in cell death. Nevertheless, recent evidence suggests alternative functions of this family of protein in metabolism regulation. In this line, we have described the involvement of the transmembrane domain of Bcl-XL in dimerization.
3- PEPCK. Together with Dr. Pascual López Buesa, we have studied several genetic polymorphisms with effect on pig meat quality, focusing in recent years on both cytosolic and mitochondrial isoforms of phosphoenolpyruvate carboxykinase. This work has evolved towards the study of post-translational regulation of this enzyme, essential for gluconeogenesis and involved in pathologies like diabetes and cancer. In a recent work, together with Dr. John Denu, from the Wisconsin Institute for Discovery, and the Universidad de Wisconsin-Madison as well as researchers from Australia, La Rioja and Harvard, we have discovered novel post-translational mechanisms of regulation of PEPCK activity in mammals. This work has been recently published in the journal Molecular Cell. We hare concluding another set of studies in this same line.
4- Parkinson´s disease. Together with Dr. Nunilo Cremades, at BIFI, we are starting a research project about Parkinson´s using a multidisciplinary approach, biophysical (Dr. Cremades) and cellular (Dr. Carrodeguas), focused on the development of novel cellular models and the use of state-of-the-art biophysical techniques with the aim of determining the initial mechanisms leading to aggregation of a-synuclein in this pathology.
5- Stem cells. We have also worked on the differentiation and death of stem cells and we are going to apply this knowledge in the development of cellular models for Parkinson´s disease.
We also collaborate with several researchers at BIFI in different research lines.
Relevant publications
- Latorre-Muro P, Baeza J, Armstrong, EA, Hurtado-Guerrero R, Corzana F, Wus LE, Sinclair DA, López-Buesa P, Carrodeguas JA, Denu JM (2018). Dynamic acetylation of cytosolic phosphoenolpyruvate carboxykinase toggles enzyme activity between gluconeogenic and anaplerotic reactions. Mol. Cell. 71: 718-732. doi: 10.1016/j.molcel.2018.07.031.
- Latorre P, Varona L, Burgos C, Carrodeguas JA, López-Buesa P. (2017). O-GlcNAcylation mediates the control of cytosolic phosphoenolpyruvate carboxykinase activity via Pgc1α. PLoS One 12: e0179988. doi: 10.1371/journal.pone.0179988.
- Hidalgo J, Latorre P, Carrodeguas JA, Velázquez-Campoy A, Sancho J, López-Buesa P. (2016). Inhibition of Pig Phosphoenolpyruvate Carboxykinase Isoenzymes by 3-Mercaptopicolinic Acid and Novel Inhibitors. PLoS One. 11: e0159002. doi: 10.1371/journal.pone.0159002.
- Escós M, Latorre P, Hidalgo J, Hurtado-Guerrero R, Carrodeguas JA, López-Buesa P. (2016). Kinetic and functional properties of human mitochondrial phosphoenolpyruvate carboxykinase. Biochem Biophys Rep. 7: 124-129. doi: 10.1016/j.bbrep.2016.06.007. Co-corresponding author.
- Latorre P, Burgos C, Hidalgo J, Varona L, Carrodeguas JA, López-Buesa P. (2016). c.A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci Rep. 6: 19617. doi: 10.1038/srep19617. Co-corresponding author.
- Nelo-Bazán MA, Latorre P, Bolado-Carrancio A, Pérez-Campo FM, Echenique-Robba P, Rodríguez-Rey JC, Carrodeguas JA. (2015). Early growth response 1 (EGR-1) is a transcriptional regulator of mitochondrial carrier homolog 1 (MTCH 1)/presenilin 1-associated protein (PSAP). Gene 578:52-62. doi: 10.1016/j.gene.2015.12.014.
- Echenique-Robba P, Nelo-Bazán MA, Carrodeguas JA. (2013). Reducing the standard deviation in multiple-assay experiments where the variation matters but the absolute value does not. PLoS One 8: e78205. doi: 10.1371/journal.pone.0078205.
- Ospina A, Lagunas-Martínez A, Pardo J, Carrodeguas JA (2011). Protein oligomerization mediated by the transmembrane carboxyl terminal domain of Bcl-XL. FEBS Lett. 585: 2935-42. doi: 10.1016/j.febslet.2011.08.012.
- Conesa C, Doss MX, Antzelevitch C, Sachinidis A, Sancho J, Carrodeguas JA (2012). Identification of specific pluripotent stem cell death–inducing small molecules by chemical screening. Stem. Cell Rev. 2012 Mar;8(1):116-27. doi: 10.1007/s12015-011-9248-4.
- Lamarca V, Marzo I, Sanz-Clemente A, Carrodeguas JA (2008). Exposure of any of two proapoptotic domains of presenilin 1-associated protein/mitochondrial carrier homolog 1 on the surface of mitochondria is sufficient for induction of apoptosis in a Bax/Bak-independent manner. Eur. J. Cell. Biol. 87: 325-34. doi: 10.1016/j.ejcb.2008.02.004.
Main research projects
1.- Modulación de las características del músculo esquelético por la fosfoenolpiruvato carboxiquinasa. Facultad de Veterinaria – Universidad de Zaragoza. Pascual López Buesa y José Alberto Carrodeguas Villar. CICYT. 2016-1018.
2.- Modulación de las características del músculo esquelético por la fosfoenolpiruvato carboxiquinasa. Instituto Universitario De Investigación De Biocomputación y Física De Sistemas Complejos – Universidad de Zaragoza. José Alberto Carrodeguas Villar. VIC. INV. – APOYO INV. 2016.
3.- PEPCK y sus efectos sobre el metabolismo, los caracteres productivos y la calidad de la carne y la canal del ganado porcino. Facultad De Veterinaria – Universidad de Zaragoza. Pascual Luis López Buesa. VIC. INV. – APOYO INV. 2015.
4.- Identificación de moléculas bioactivas en células troncales mediante cribado funcional de quimiotecas: herramientas para terapias seguras. Instituto de Biocomputación y Física de Sistemas Complejos. José Alberto Carrodeguas Villar. Universidad de Zaragoza/Ibercaja. 2012-2013.
5.- Proteínas Mtch: regulación transcripcional en humanos y efectos fenotípicos del mutante en Drosophila. Instituto Universitario De Investigación De Biocomputación y Física De Sistemas Complejos. José Alberto Carrodeguas Villar. Universidad de Zaragoza. 2011.
6.- Genética química para la identificación de compuestos bioactivos que promueven diferenciación específica, proliferación o apoptosis en células madre. Instituto de Biocomputación y Física de Sistemas Complejos. Departamento de Bioquímica y Biología Molecular y Celular. Facultad de Ciencias. Universidad de Zaragoza. José Alberto Carrodeguas Villar. Instituto Aragonés de Ciencias de la Salud. 2011.
7.- Regulación de la actividad de proteínas proapoptóticas mitocondriales. Instituto de Biocomputación y Física de Sistemas Complejos. Departamento de Bioquímica y Biología Molecular y Celular. Facultad de Ciencias. Universidad de Zaragoza. José Alberto Carrodeguas Villar. Ministerio de Ciencia e Innovación. 2010.
8.- Identificación de compuestos químicos que inducen diferenciación celular específica o muerte celular apoptótica en células madre embrionarias de ratón (continuación). Instituto de Biocomputación y Física de Sistemas Complejos. Departamento de Bioquímica y Biología Molecular y Celular. Facultad de Ciencias. José Alberto Carrodeguas Villar. Instituto Aragonés de Ciencias de la Salud. 2009-2010.
9.- Identificación de compuestos químicos que inducen diferenciación celular específica o muerte celular apoptótica en células madre embrionarias de ratón (continuación). Instituto de Biocomputación y Física de Sistemas Complejos. Departamento de Bioquímica y Biología Molecular y Celular. Facultad de Ciencias. José Alberto Carrodeguas Villar. Instituto Aragonés de Ciencias de la Salud. 2008-2010.
10.- Mecanismos moleculares de proteínas de la membrana externa mitocondrial similares a transportadores implicadas en apoptosis. Papel en enfermedades degenerativas y en cáncer. Facultad de Veterinaria. Universidad de Zaragoza. José Alberto Carrodeguas Villar. MEC. 2006-2009.
Collaborators
- Miguel Fernández Moreno and Juan José Arredondo. Instituto de Investigaciones Biomédicas. CSIC-UAM. Madrid.
- Javier Sancho, Milagros Medina, Adrián Velázquez Campoy, Ramón Hurtado-Guerrero, Patricio Fernández Silva, Raquel Moreno Loshuertos. Instituto de Biocomputación y Física de Sistemas Complejos. Departamento de Bioquímica y Biología Molecular y Celular. Facultad de Ciencias. Universidad de Zaragoza.
- José Carlos Rodríguez-Rey. Department of Molecular Biology, University of Cantabria, IDIVAL, Santander, Cantabria, Spain.
- Flor Pérez Campo. Department of Internal Medicine, Hospital U. Marqués de Valdecilla-IDIVAL University of Cantabria, 39008 Santander, Cantabria, Spain.
- John M. . Wisconsin Institute for Discovery, Morgridge Institute for Research, and the Department of 12 Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, USA.
- Francisco Corzana. Departamento de Química, Centro de Investigación en Síntesis Química, Universidad de La Rioja.
Computational Genomics and Systems Bio-medicine
Head of the Research Line:
Joaquín Sanz
Researchers:
PhD students:
Mario Tovar
Jorge Cárdenas
Master students:
Ignacio Marchante
Undergrad students:
Santiago Royo
Pablo Pérez
Pilar Cobos.
Alumni:
Jessica Moreira Batista Da Silva
Regina Santesteban Azanza
SUMMARY OF THE RESEARCH LINE
In our lab, we use mathematical models to describe infectious and auto-immune diseases at a variety of scales: from cells & genes to individuals & populations. Our main goal is to pinpoint the causal factors -both genetic and environmental- shaping variation in immune responses to pathogens, as well as to characterize the drivers of functional vs. pathological responses and learn how do they relate to epidemiological observations.
To do that, NGS genomic data constitutes our main raw material. That includes genetic variation data on large human cohorts, as well as host & pathogen transcriptomes and epigenomes, both in human & animal models at bulk and single-cell resolution. On the epidemiological side, we develop transmission models to interpret data such as trans-national burden registers, prevalence surveys and clinical trials outcomes. In pursuing these goals, our methods integrate tools from computational genomics, systems Bio-Medicine, biostatistics, data & network science, Bayesian inference, mathematical epidemiology and Physics of complex systems.
Nowadays, there is three main research thread open in the line, which was created in 2019. These include the Systems Biology of host-pathogen interactions in tuberculosis; the development of computational tools for single-cell -omics data analysis, and the development of computational applications to the study of the genomics of the immune system.
Systems biology of tuberculosis.
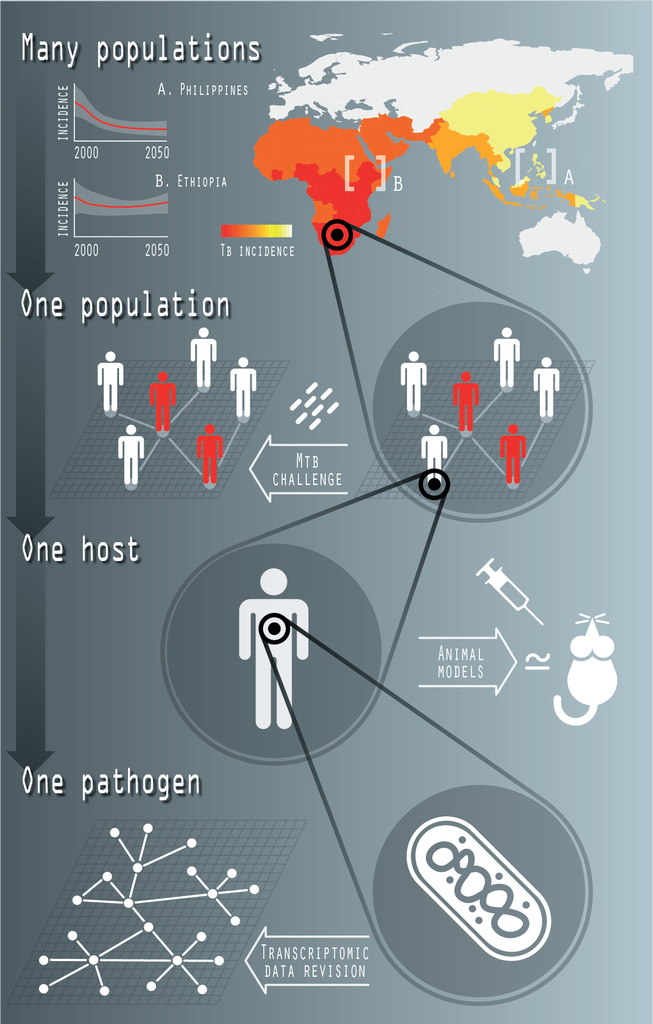
Tuberculosis (TB) is one of the most ancient infectious diseases affecting humans, and, with an estimated number of 1.4 million deaths in 2019, it remains among the deadliest ones. Its causative agent, the bacillus Mycobacterium tuberculosis, is arguably the most successful among all human pathogens, considering its striking ability to coexist with its host (circa. 24% of contemporary humans are estimated to be infected with M.tb.) without compromising their fitness. Among all possible epidemiological interventions under consideration in the fight against TB, the introduction of a new vaccine able to complement, or outperform the current vaccine of the Bacillus Calmette-Guerin (BCG) holds the promise of a greatest impact against the disease.
In our lab, we use Systems Biology, Bio-informatics & Mathematical Epidemiology techniques to model Tuberculosis infection and TB vaccine properties at different levels of complexity. That includes the genomic characterization of the regulatory mechanisms underlying mycobacterial-induced trained innate immunity, and the epidemiological modelling of TB transmission at a trans-national level to evaluate the impact of epidemiological interventions and assist the analysis and interpretation of vaccine-efficacy clinical trials.
Fig. 1: Systems Biology of TB. The recent activity of the group in this topic includes the characterization of transcriptional regulatory networks, at the level of single bacteria; the study of innate trained immunity in collaboration with M. Divangahi (McGill) and his team (at the level of one host); the ongoing characterization of the genetic architecture of the response of human macrophages to M.tb. infection (at the level of one population), and the development of mathematical models of TB transmission, at supranational scales (many populations).
Single-cell -omics data modelling and analysis
In 2009, Fuchou Tang and collaborators were the first to use single cell RNA-seq, from a four-cell stage mouse blastomere. Since then, the field has witnessed spectacular progress in microfluidic techniques, automation of library preparation and multiplexing, to the point that now it is possible, and relatively affordable, to compile datasets containing hundreds of thousands of cells. These experimental improvements unlock the possibility of addressing deeper Biological problems through the usage of complex experimental designs.
In the lab, we combine data-science, statistical modeling and complex networks methods to propose improved pipelines for single cell transcriptomic data, with a special focus on characterizing transcriptional responses to infection, immune training, and vaccination in immune cells and their hematopoietic precursors.
Computational immuno-genomics
Immune responses to immunological insults are complex, and highly variable across individuals. The adequacy of the immune response to an infection challenge can be often evaluated by observing its strength, promptness and specificity; and all these are dynamical features that emerge from a complex interplay between many different causal factors. These factors equally stem from the genetics of both host and pathogen as well as from the environmental context wherein they meet.
In the lab we study the causality links that connect genotypes and environmental variables with immune responses to infection. Our main objective is the identification of the components of such responses that are more strongly affected by those causal factors, as well as their evolutionary and clinical implications.
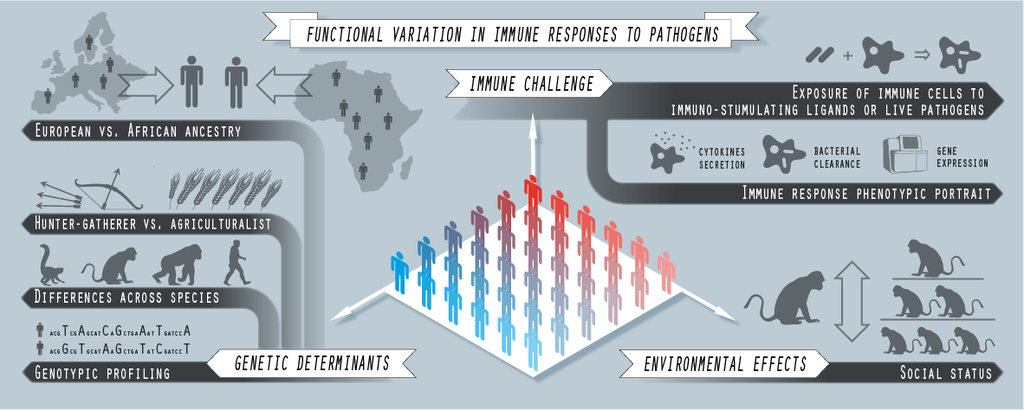 Fig. 2. Computational immunogenomics. Graphical scheme of the experimental approaches implemented in some of the last projects we have been reciently involved with, in collaboration with professors L. Barreiro (U. Chicago), and J. Tung (Duke), and their teams.
Fig. 2. Computational immunogenomics. Graphical scheme of the experimental approaches implemented in some of the last projects we have been reciently involved with, in collaboration with professors L. Barreiro (U. Chicago), and J. Tung (Duke), and their teams.
Relevant publications
- Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity. Hawash, M., Sanz, J, Grenier, J. C., Kohn, J., Yotova, V., Johnson, Z., … & Barreiro, L. B. (2020). Proceedings of the National Academy of Sciences, 2021.
- tuberculosis Reprograms Hematopoietic Stem Cells to Limit Myelopoiesis and Impair Trained Immunity. Khan1, Downey, Sanz, J., Kaufmann, Blankenhaus, Pacis, … & Divangahi (2020). Cell, 183(3), 752-770.
- Social history and exposure to pathogen signals modulate social status effects on gene regulation in rhesus macaques. Sanz, J., Maurizio, P. L., Snyder-Mackler, N., Simons, N. D., Voyles, T., Kohn, J., … & Barreiro, L. B. (2020). Proceedings of the National Academy of Sciences, 117(38), 23317-23322.
- Bridging the gap between efficacy trials and model-based impact evaluation for new tuberculosis vaccines. Tovar, M., Arregui, S., Marinova, D., Martín, C., Sanz, J., & Moreno, Y. Nature Communications, 10(1), 1-10. (1 co-last author) (2019).
- Natural selection contributed to immunological differences between hunter-gatherers and agriculturalists. Harrison, G.F., Sanz, J., Boulais, J., Mina, M.J., Grenier, J.C., Leng, Y., Dumaine, A., Yotova, V., Bergey, C. M., Nsobya, S.L., Elledge, S.J., Schurr, E., Quintana-Murci, L., Perry, G.H., Barreiro, L.B. Nature Ecology and Evolution 1253-1264 3(8) (2019).
- Spotting the old foe—revisiting the case definition for TB. Houben, R.M., Esmail, H., Emery, J.C., Joslyn, L.R., McQuaid, C.F., Menzies, N.A., Sanz, J., Shrestha, S., White, R.G., Yang, C. and Cobelens, F., 2019. The Lancet Respiratory Medicine, 7(3), pp.199-201.
- Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques. Snyder-Mackler, N., Sanz, J., Kohn, J. N., Voyles, T., Pique-Regi, R., Wilson, M. E., … & Tung, J. (2019). Proceedings of the National Academy of Sciences, 116(4), 1219-1228.
- Genetic and evolutionary determinants of human population variation in immune responses. Sanz, J., Randolph, H. E., & Barreiro, L. B. (2018). Current opinion in genetics & development, 53, 28-35.
- Data-driven model for the assessment of Mycobacterium tuberculosis transmission in evolving demographic structures. Arregui, S., Iglesias, M. J., Samper, S., Marinova, D., Martin, C., Sanz, J. , & Moreno, Y. 1 . (1 co-last author). (2018). Proceedings of the National Academy of Sciences, 115(14), E3238-E3245.
- BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Kaufmann, E. 1, Sanz, J. 1, Dunn, J. L. 1, Khan, N., Mendonça, L. E., Pacis, A., … & Mailhot-Léonard, F. (1 co-first author). (2018). Cell, 172(1-2), 176-190.
- Social status alters immune regulation and response to infection in macaques. N. Snyder-Mackler1, J. Sanz1, J.N. Kohn, J.F. Brinkworth, S. Morrow, A.O. Shaver, J.C. Grenier, R. Pique-Regi, Z.P. Johnson, M.E. Wilson, L.B. Barreiro2 & J. Tung2 (1 co-first author; 2 co-last author); Science, 354 (6315), 1041-1045.
- Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Y. Nédéléc1, J. Sanz1, G. Baharian1, Z.A. Szpiech, A. Pacis, A. Dumaine, J.C. Grenier, A. Freiman, J. Sams, S. Hebert, A. Pagé-Sabourin, F. Luca, R. Blekhman, R.D. Hernández, R. Piqué-Regi, J. Tung, V. Yotova & L.B.B. Barreiro, (1 co-first author). Cell 167-3, p657–669.e21 (2016) (The paper was selected for the Issue cover).
Main research projects
- PID2019-106859GA-I00: Enfoques sistémicos a los mecanismos de defensa del hospedador ante enfermedad e infección en M. tuberculosis: causas genéticas y evaluación de impacto en nuevas vacunas, 2020-2023 Spanish Ministry of Science and Innovation (MICINN), Principal Investigator: Joaquín Sanz.
- Bio-computational approaches applied to the development of TB vaccines: epidemiological modeling, efficacy simulations and immunogenetics analyses. Government of Aragon, Spain. Grant LMP117-18. 2019-2020. PI: Yamir Moreno.
- Multi-scale approaches to Tuberculosis infection: mathematical epidemiology and functional genomics. National Programme for Recruitment and Incorporation of Human Resources 2018, subprogramme “Ramón y Cajal”. RYC-2017-23560. 2019-2024. Principal Investigator: Joaquín Sanz.
- Stress and the Genome: Testing the Impact of Social Effects on Gene Regulation. National Institute of Health NIH (USA) Project # 1R01GM102562-01. (2012-2022). Principal Investigator: Dr. Jenny Tung. (Duke University)
Collaborators
- Maziar Divangahi, McGill University, Canada.
- Eva Kaufmann, McGill University, Canada.
- Luis Barreiro, University of Chicago, USA.
- Genelle Harrison, University of Colorado, USA.
- Jenny Tung, Duke University, Durham, USA.
- Bana Jabri, University of Chicago, USA.
- Valentina Discepolo, Universitá Federico II, Naples, Italy.
- Carlos Martín, unizar.
- Nacho Aguiló, unizar.
- Jesús Gonzalo-Asensio, unizar.
- Yamir Moreno, unizar.
- Pierpaolo Bruscolini, unizar.
- Mario Floría, unizar.
- Jesús Gómez-Gardeñes, unizar.
- Sandro Meloni, IFISC, Palma de Mallorca.
Contact
Jsanz@bifi.es
https://sanzlab.wordpress.com/